 email Project Administrator
email Project AdministratorJoin this project
 email Project Administrator
email Project Administrator
Join this project
| The Y-Haplogroup J2b_455-8 Project
| 
|
The value 8 repeats on Short Tandem Repeat (STR) marker DYS455 (Family Tree DNA marker #16) is very rare. There is a group of a few Ashkenazim who are in Haplogroup J2b2 with fairly closely matching Y-STR profiles. They have a rare deletion event of three repeats in DYS455 which exist so far only in one other haplogroup (I1a). They also have the unusual value of 19-20 on YCAii. The International Society of Genetic Genealogy (ISGG) called at one time their haplogroup subclade as J2b2e. However, since then they stop using that subclade name because they are now using only single-nucleotide polymorphism (SNP) in defining their haplogroup subclades and the e was based on a STR.
Family Tree DNA also had change the way they call their Y-DNA
haplogroup subclades. They are now naming them the major haplogroup
following by the latest positive SNP that was tested.
The way they used to define them
was to add a number or letter for new each newer SNP mutation
they found. This however in some haplogroups produced a
series of characters 22 characters or longer. Therefore
members of this projects would have several different
haplogroups listing based on which SNPs they were tested
on. Below is the list of old haplogroup names with the new ones with Years Before Present (ybp) estimated by Yfull.com.
| Old Name | Family Tree DNA | YFull | Formed Estimate | TMRCA | |||||
|---|---|---|---|---|---|---|---|---|---|
| J | J-M304 | J-M304f | 42800 ybp | 31300 ybp | |||||
| J2 | J-M172 | J-M172 | 31300 ybp | 27700 ybp | |||||
| J2b | J-M102 or J-M12 | J-M102 | 27700 ybp | 18100 ybp | |||||
| J2b2~ | J-Z529 | J-Z534 | 15900 ybp | 15900 ybp | |||||
| J-Z1827 | J-Z1825 | 15900 ybp | 13900 ybp | ||||||
| J-Z593 | J-Z593 | 13900 ybp | 12800 ybp | ||||||
| J2b2a | J-M241 | J-M241 | 12800 ybp | 5900 ybp | |||||
| J2b2a1 | J-L283 | J-L283 | 9700 ybp | 5700 ybp | |||||
| J-Z622 | J-Z622 | 5700 ybp | 5400 ybp | ||||||
| J2b2a1a~ | J-Z600 | J-Z600 | 5400 ybp | 5400 ybp | |||||
| J2b2a1a1~ | J-Z628 | J-Z585 | 5400 ybp | 5400 ybp | |||||
| J2b2a1a1a~ | J-Z615 | J-Z615 | 5400 ybp | 4700 ybp | |||||
| J-Z597 | J-Z597 | 4700 ybp | 4400 ybp | ||||||
| J2b2a1a1a1~ | J-Z2507 | J-Z2507 | 4400 ybp | 4400 ybp | |||||
| J-Z1246 | |||||||||
| J2b2a1a1a1b~ | J-CTS3617 | J-Y15058 | 4400 ybp | 4000 ybp | |||||
| J-Z38240 | J-Z38240 | 4000 ybp | 3900 ybp | ||||||
| J2b2a1a1a1b3~ | J-CTS6190 | J-CTS6190 | 3900 ybp | 3300 ybp | |||||
| J-Z34474 | |||||||||
| J-Y34371 | J-Y34371 | 3300 ybp | 3300 ybp | ||||||
| J2b2a1a1a1b3b~ | J-Y33799 | J-Y33795 | 3300 ybp | 1800 ybp |
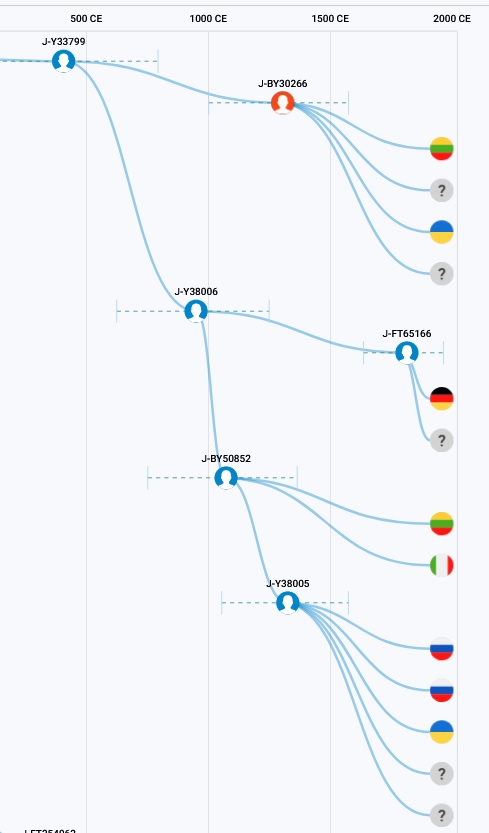
If we all have been tested on SNP Y33799 only, we would all be listed now as Y33799 at Family Tree DNA. This is because the defining mutation for our subclade is DYS455 becoming the 8 happened around 900 ybp after all the above SNPs. Therefore there is no reason to order any advanced SNP Testing for any of the above SNP since all fifteen members that were tested at Family Tree DNA with the Big-Y test have the Y33799 SNP which occurred before 900 years ago. Seven of the ten had SNP BY50852, while the other three have SNP BY30266. About half the persons in the project have a rare mutation on marker DYS464 which occurred about 500 years ago after the BY30266 mutation since both persons having that marker mutation that were tested have that SNP mutation along with someone that did not have it. If you were tested on the Family Finder test and not the Big-Y, you will be listed as J-CTS6190.
Other testing companies are using other naming method and one may see us listed as J2b2* or maybe even J2b2a1a1b1a. 23andMe now list us at J-L283 However some people listed as such will have 11 and not 8 on DYS455 because these companies also are using only SNPs and the * means there could be other SNP mutations that they did not tested for.
Dr. Whit Athey in his article about our sub-haplogroup in the "Journal of Genetic Genealogy", said that our haplogroup is less than 1000 year old and was Ashkenazims. (You can watch him on the 2009 television show "Tracing Your Family Roots" talked about our group.) Using the STRs values of all members that were tested on as least 25 markers the estimate now is about 900 years ago.
We presently have 151 different surnames from the 230 people with surnames. The reason for this is that most Ashkenazims did not take surnames until the late 1700s and early 1800s. Before then, they were known are "name son of name". The people known to be a J2 with DYS455= 8 came from eight difference sources: members of this Family Tree DNA Project (171), others tested at Family Tree DNA (69), Y-search (13), Sorenson Molecular Genealogy Foundation (SMGF) (3), Ancestry.com (13), DNA Heritage, Dr. Athey article (2), and Dr. Hammer and others' sample of Jewish priesthood (15).
 All but one family of the project have a family lore
of being an Kohanim. In a recent Jewish priesthood
study by Dr. Michael F. Hammer with others, we
were 6% of all the Kohens. Including the other J2 Kohens, the
total were 43% of the Kohanim. When members of our group
asked Bennett Greenspan, President of FamilyTreeDNA about
this, his answer was
All but one family of the project have a family lore
of being an Kohanim. In a recent Jewish priesthood
study by Dr. Michael F. Hammer with others, we
were 6% of all the Kohens. Including the other J2 Kohens, the
total were 43% of the Kohanim. When members of our group
asked Bennett Greenspan, President of FamilyTreeDNA about
this, his answer was
|
The CMH appears to be in J1 instead of
J2...however there is a minor subgroup in
J2 that has the oral tradition of being
Cohanim. As you probably know the
historian Josephus says that during the
later Temple II period the position of
Cohanim Godal became corrupted with
people with $$$ purchasing the position,
but we can't ever be 100% sure whether
the major group in J1 or the minor group
in J2 is the real "McCoy" so to speak. |
Members of this project only can look at the Y-DNA STR table of the members. The Family Tree DNS's haplogroup project is divided into 5 groups.
| Group No. | DYS 393 | DYS 464 | CDY | TMCRA |
|---|---|---|---|---|
| 1 | 12 | 13-14-15-18 | 35-39 | 900 years |
| 2 | 13 | 13-14-15-18 | 35-39 | 600 years |
| 3 | 13 | 13-13-14-15 | 35-39 | 500 years |
| 4 | 13 | 13-13-14-15 | 35-38 | ?? years |
| 5 | 13 | 13-13-14-15 | ?? | 500 years |
| 6 | 13 | ?? | ?? | 600 years |
So far, everyone with DYS393 value of 13 with test result of DYS455 have 8 on the later. Therefore the chances that everyone that match one of the project members with only 12 markers and with DYS393 = 13 also are welcome in the project.
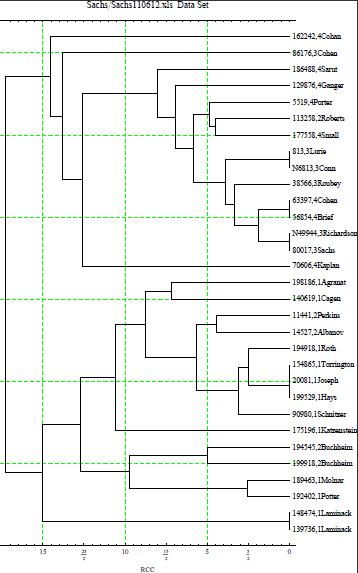
William E. Howard III has developed a method that use
correlations for the analysis of pairs of 37 markers Y-STR
haplotypes which he named RCC. Each RCC is roughly equal to
a little less than 50 years. He then used a computer program
called Mathematica to make the tree which is shown below.
Because of randomness of mutations, the pairing could be off by
up to 300 years. The way you can estimate when the MRCA
between any two persons lived, find the RCC at the junction
point connecting the two and multiply it by 50. This would give
you an time estimate within 300 years. RCC values when paired
have a standard deviation of about 3, or 150+ years, but the
RCCs of the junction points probably are slightly more
trustworthy.
We also have one person with another type of STR mutation. This person have 6 copies of DYS464 instead of the usual 4 copies.
Click here to join this project
Note: Method how the Mutations were dated which is based on the mutation rates of John F. Chandler as listed in Journal of Genetic Genealogy 2:27-33, 2006. Other people may use other mutation rates. For example, the Heinila STR-mutation rates will give an earlier date by about 40 percent. Also the years per generation that I used was 28.8 which may be too low.