A New Subclade of Y Haplogroup J2b
T. Whit
Athey and Bonnie E. Schrack
Abstract
Evidence
from case studies is presented that allows the definition of a new subclade of
Y Haplogroup J2b-M102. The defining
binary polymorphism of the new subclade is a deletion event in the Y-STR marker, DYS455. The present
study shows that the new subclade is located in the J2b phylogeny downstream
from the previously defined subclade J2b-M241. The members of the subclade were
asked about Ashkenazi and Cohanim backgrounds on their patrilineal lines and of
the four who responded, all four are Ashkenazi Jews and Cohanim. This suggests that future Y-chromosome
studies involving Ashkenazi and Cohanim should type DYS455 to investigate further this possible link.
Address for
correspondence: Whit Athey, wathey@hprg.com
Received: January 9, 2008; accepted: February 27,
2008
Introduction
About
one-fifth of Ashkenazi Jews are members of Y-Haplogroup J2 (Behar, 2004). Previous studies of Ashkenazim did not employ
sufficient resolution to understand exactly where the Jewish groups were
located in the J2 phylogeny, or if there were any subclades of J2 that were
almost exclusively Jewish. The present
study focuses on one small part of Haplogroup J2 and demonstrates that a new
subclade of Haplogroup J2b-M12 may be defined.
All members of the subclade who have been identified so far are
Ashkenazi and Cohanim, though the sample is small.
Figure
1 provides a
simplified overview of Haplogroup J2 (ISOGG, 2007) and shows the location and
phylogenetic structure of Haplogroup J2b.

Figure
1. Simplified phylogenetic chart for Haplogroup J2
according to ISOGG (2007). Text in red
font indicates defining binary polymorphisms.
It is probable
that the binary polymorphism (BP) M102 may be redundant with M12, but previous
publications of Haplogroup J phylogenetic trees, including that of ISOGG
(2007), still show these BPs as reflected in Figure 1. It is likely that there will be other changes
to the 2008 version of the ISOGG tree, necessitated by the publication of new
results during the past year.
Within
Haplogroup J2b1-M102, there are presently two subclades, J2b1a defined by M241
and J2b1b defined by M205 (ISOGG, 2007).
Note that some phylogenetic trees have the position of M241 and M205
interchanged, so in this article, the defining BP will be used to make it clear
which subgroup is intended. On the basis
of limited data, J2b1a-M241 appears to be the larger of the two subgroups
(Sengupta 2006). Several customers of
Family Tree DNA (FTDNA, Houston, TX) who are M102+ (in J2b1) have the unusual value of
8 on the Y-STR marker, DYS455.
The Y-STR marker DYS455 consists of a poly-(AAAT)
sequence with the great majority of men having a repeat value of 11. A repeat value of 8 on DYS455 has previously been observed
only in members of Haplogroup I1a, where the value resulted from a three-repeat
deletion event that occurred close to the founding event for Haplogroup I1a,
because thus far no members of I1a have failed to exhibit the deletion
event. The deletion is stable within
Haplogroup I1a with only about 2% of members having mutated to values of 7 or 9
in the 10,000+ year history of the haplogroup.
Those J2b
members with DYS455=8 have apparently also inherited this value from a common ancestor
who, given the similarity in their Y-STR profiles, probably lived less
than one thousand years ago.
Because DYS455=8 occurs only in Haplogroup
I1a and in the J2b subclade that we are presently considering, it is quite
probable that the three-repeat deletion event in DYS455 has occurred just twice in
human history. Alternately, if it has
occurred more often, only two of the founders have living descendants. In the present study we show that the
deletion event in a J2b founder has allowed us to define a new subclade within
Haplogroup J2b. Testing of binary
polymorphisms (BPs) in several subjects demonstrates that the new subclade is
located within Haplogroup J2b-M241, though the exact relationship to the other
subgroups of J2b-M241 is still uncertain, as discussed in more detail below. We have named the new subclade, J2b1a4,
though revisions to the J phylogenetic tree will likely cause the name to
change.
Methods
Four
subjects were recruited who had previously been tested and found to be M205+,
along with four subjects who had previously been tested as M241+. Two of these eight subjects had been tested
on both M241 and M205. Four subjects who
were M102+ and who had DYS455=8 were also recruited.
Two of the latter two subjects had also previously been tested on M205
at FTDNA and had been found to be negative.
These two subjects were then tested by Ethnoancestry for M241. One of these has also been tested on two
SNPs, M99 and M280, that define subgroups of J2b-M241. Four other subjects with DYS455=8, but who have no BP results,
were also included in the study for comparison on Y-STR values. These twelve participants in the present
study were designated as Subjects 101-112.
The
public databases of the Sorenson Molecular Genealogy Foundation (SMGF),
Y-Search, and Y-Base were searched for occurrences of DYS455=8. Those four that also appeared to be members
of Haplogroup J2b, according to the haplogroup prediction program of Athey
(2005, 2006), were included in the study as Subjects 113-116, in order to
further characterize the Y-STR values of the new subclade.
For the
present study, testing of M241 was carried out on selected subjects by
Ethnoancestry (Cyprus, CA). Testing of the BPs M99 and M280 was carried
out previously at FTDNA. At Ethnoancestry,
Y chromosome BPs are amplified by PCR with standard primers giving
products from 200 to 500 bp in length. PCR products are then sequenced using
dye terminator chemistry with electrophoresis on a capillary ABI sequencer. Alleles are called in the software package,
Sequencher, by alignment with chromosomes of known allelic state (positive and
negative controls). The procedures
followed at FTDNA have not been published, but they are probably similar.
Results
The
results for each subject on several BPs are shown in Table 1. These results can be used to locate the DYS455 deletion event within the
phylogeny of Haplogroup J2b. Because the
detailed logic model for placing a new BP within an existing two-branch /phylogeny
is not usually included in research articles, it will be discussed in some
detail here. Following the discussion of
the placement of the new subclade, this article will also discuss in some
detail, the issue of when it is appropriate to use a deletion in a Y-STR marker for phylogenetic purposes,
and then apply the resulting principles to the present case.

In the
following discussion and in Table 1, the BP representing the deletion
event in DYS455 will be designated as “455del.”
A person who has a value of 8 on DYS455 (and rarely 7 or 9) will be
termed 455del+, while a person who has the value of 11 (or rarely 10 or 12)
will be termed 455del-.
Placement
of the New Subclade Within J2b1
All of
the subjects in the present study who have DYS455=8 and test results for M102
were found to be M102+. Therefore, the
placement of the new clade focused on the structure within J2b1-M102. Figure 1 shows that the structure of
J2b1-M102 consists of two subgroups defined by M241 and M205. In the following, we assume that this basic
structure is correct—that neither M205 nor M241 is upstream or downstream from
the other, and that they define parallel branches as shown. The new subclade defined by DYS455=8 cannot be parallel to the
other two branches since at least two of its members are also M241+.
Therefore,
we now develop a logic model for placing a new subclade within this or a
similar structure. The approach is to
lay out all of the possible placements for the BP 455del. In most cases, only a single counter example
is necessary to disprove a potential placement, though a second confirming case
is helpful in ruling out lab errors. If
all possible placements except for one can be disproved, then the remaining one
is the correct one. Looking only for
positive evidence for the most likely placement would require a large number of
subjects to provide reasonable assurance of the placement being correct. The present approach is efficient in that
only a small number of well-chosen subjects will suffice to place the new
subclade.
a. In the following discussion, Figure 2
outlines all of the possible placements of the new subclade of J2b1 defined by
455del. We will consider each
possibility in turn, and show what would be necessary to negate or disprove
each possibility. The possibility remaining
after this process of elimination, the one that cannot be rejected, is the
correct placement.
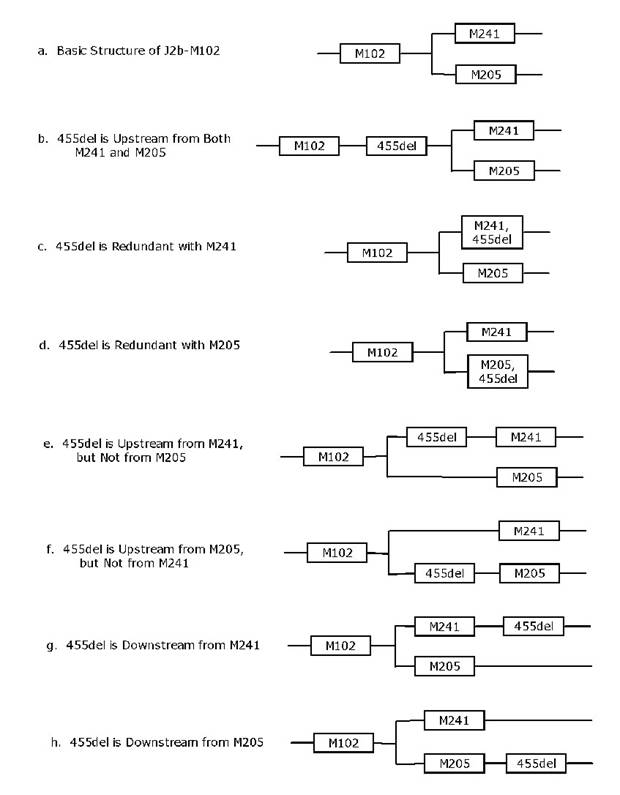
Figure 2. The seven
possible phylogenetic locations for the deletion event in DYS455 with respect to M102, M241,
and M205.
b. The 455del is upstream from both M241 and
M205 as shown in Figure 2b.
In this
case, all M241+ and M205+ subjects will also have 455del. However, if any one M241+ or M205+ subject
does not have the 455del, then this possibility is rejected. In this study subjects 101-104 are M241+ and
105-108 are M205+ and all have 455del- (e.g., they do not have the
deletion). Therefore, this possibility
is rejected.
c. 455del is redundant with M241 as shown in Figure
2c.
To
disprove this possibility, we need only to find at least one 455del+ subject
who has M241-, or at least one M241+ who has 455del-. In the present study, subjects 101-104 are
all M241+, but none has the deletion.
Therefore, this possibility is rejected.
d. 455del is redundant with M205 as shown in Figure
2d.
To disprove
this possibility, we need only to find at least one 455del+ subject who has
M205-, or at least one M205+ subject who has 455del-. In the present study, subjects 105-108 are
all M205+, but none has the deletion.
Therefore, this possibility is rejected.
e. 455del is upstream on the branch leading to
M241 as shown in Figure 2e.
To
disprove this possibility, we need only find at least one M241+ subject who is
negative for 455del. This disproof is
provided by subjects 101-104, all of whom are M241+ and 455del-.
f. 455del is upstream on the branch leading to
M205 as shown in Figure 2f.
To
disprove this possibility, we need only show that there exists at least one
M205+ subject who is negative for 455del.
This disproof is provided by subjects 105-108, all of whom are M205+ and
455del-.
g. 455del is downstream from M241 as shown in Figure
2g.
To
disprove this possibility, we would need to find a subject who exhibits the
deletion in DYS455, but is M241-. We found no
such results, so this remains an open possibility.
h. 455del is downstream from M205 as shown in Figure
2h.
To
disprove this possibility, we would need to find a subject who exhibits the
deletion in DYS455, but is M205-. Subjects 109
and 112 have 455del+, but are M205-, so they provide this disproof.
Summarizing,
we have shown that every possibility except for (g) has been disproved. Since one possibility must be true, it must
be (g). Indeed, subjects 109 and 112
have results that are consistent with 455del being downstream from M241.
Placement
of the New Subclade Within J2b1a-M241
Within
Haplogroup J2b1a-M241, there are already three previously known subgroups
(ISOGG 2007), so the subclade defined by 455del could be (1) a fourth subgroup
in parallel to the existing three subgroups, (2) downstream from one of the
three, (3) upstream from one or more of the three, or (4) redundant with one of
the three. In principle, we could use
the same kind of logic model and apply it to the placement of 455del within
J2b1a-M241. However, at the present
time, only two BPs, M99 and M280, out of the three defining these three
subgroups of J2b1a-M241, have commercially available tests, so the exact
placement of the new subclade cannot be completely resolved at this time. Subjects 109 and 112, who have DYS455=8, were found to be negative
for M99 and M280, so we can at least be sure that 455del is not downstream from
either of these two SNPs. In other words,
the subclade defined by the deletion is not a subgroup of J2b1a1-M99 or
J2b1a2-M280. We were unable to identify
any subject who was M99+ or M280+, so we cannot say with certainty that one of
these two BPs is not downstream from the deletion event. Tentatively, the new subclade can be placed
as a parallel branch to the three other subgroups of J2b1a-M241 as shown in Figure
3.

Figure
3 Haplogroup J2b1 with the new subclade shown
(simplified for clarity).
The
Use of DYS455=8 for Phylogenetic Purposes
If the
deletion in DYS455 occurred in the region flanking the (AAAT)n repeat
structure, then it would be considered just another a binary polymorphism like
many other indels included on the Y phylogenetic tree. However, the PCR product for DYS455 has not been sequenced to show
the exact nature of the deletion event, so we must assume the worst case—that
the deletion occurred entirely within the poly-(AAAT) structure, so that the
remaining part of the repeat structure would continue to behave as a normal,
albeit truncated, Y-STR marker. Therefore,
we must examine the appropriateness of using this particular deletion in a Y-STR marker for phylogenetic purposes.
To be
appropriate for use for phylogenetic purposes, the Y-STR marker should have the following
characteristics:
Criterion
1. The marker should be quite stable at
its new value, with essentially no chance of reverting to its previous value of
11.
Criterion
2. Further mutations in the marker
should not produce an allele frequency distribution that overlaps the former
distribution.
Criterion
3. The deletion event should be “almost
unique,” perhaps occurring no more than a few other times in human
history. There are several BPs on the Y
phylogenetic tree that have occurred two or more times in different parts of
the tree. Where BPs are not unique, they
must be easily interpretable within the context of the phylogeny. Absolute uniqueness has never been required
of a marker on the Y tree.
A final
characteristic should be mentioned, though it applies to other types of markers
as well—the marker should be useful for phylogenetic purposes. For example, there is no reason to use the
deletion in DYS455 within Haplogroup I because there are already a number of
redundant SNPs available to define Haplogroup I1a. The deletion in DYS455 is useful within Haplogroup
J2b because there is apparently not currently a SNP available to define the
subclade.
Starting
with the last criterion, the value DYS455=8 exists also in Haplogroup
I1a, but it has not been reported in any other haplogroup. Therefore, the “almost unique” criterion is
met. Since the modal value within
Haplogroup I is also 11, it would appear that a similar three-repeat deletion
has occurred, though the marker has not been sequenced in I1a either. Therefore, we can examine the stability
criterion within Haplogroup I1a where a very large amount of data is available.
A group
of 168 haplotypes that had been tested and found positive for one of the
defining BPs for Haplogroup I1a (usually M253) was extracted from public
surname project web pages at the FTDNA web site. Duplicates from the same surname cluster were
not included. 166 of the haplotypes had
a value of 8 on DYS455, while two had a value of 9. Another group of 374 haplotypes were
extracted from Y-Search where Haplogroup I1a was indicated as the haplogroup,
again not counting duplicate surnames, and the DYS455 values for these were also
examined. Only nine of the 374
haplotypes had values different from 8—two haplotypes had values of 7 and seven
had values of 9. Therefore, since we see
only a very sharp distribution centered at DYS455=8, in spite of Haplogroup I1a
likely being more than 10,000 years old (Rootsi 2004), the marker at a repeat
value of eight appears to be remarkably stable.
In Haplogroup I1b-S31, DYS455=11 is the modal value, and out
of over 300 haplotypes extracted from Y-Search, only one had a value of ten,
and one had a value of 12. Therefore,
the allele frequency distribution on DYS455 for the remainder of
Haplogroup I does not overlap the data from I1a. However, if we examined a sample of millions
of people, there could a small degree of overlap.
Haplogroup
J is older than I1a, so we would expect to see a broader distribution of repeat
values on DYS455. As much as approximately
5% of Haplogroup J2a has a value of 10 on DYS455, but no values of 9 have yet
been reported. Among a group of 100
people who were tested or predicted by FTDNA to be in J2b, none had the value
10 and only one had a value of 12.
With only
a small number of members of Haplogroup J2b-DYS455=8 being thus far identified,
it is not surprising that the only repeat value on DYS455 that has been observed is
eight. It can be expected that the
allele frequency distribution on DYS455 will be similarly stable and
no less sharply distributed about the value of 8 as was found in Haplogroup
I1a.
Therefore,
all three criteria are satisfied and the deletion event in DYS455 may be used for phylogenetic
purposes within Haplogroup J2b.
It should
be noted that this is not the first use of a Y-STR marker for phylogenetic purposes
(DiGiacomo 2004; Sengupta 2006, ISOGG 2007, King 2008).
Origins
of the Subclade
There are
currently 10 people in the database of FTDNA who have DYS455=8 and also appear to be
members of Haplogroup J2b, either through testing of M12 or prediction based
upon Y-STR haplotype. Three others appear to belong to the same Y-STR cluster, but they have not tested
DYS455.
All of
those members of J2b-DYS455=8 who have entries on Y-Search, including Subjects
109, 110, and 112, and who identify their patrilineal family background,
indicate Ashkenazi Jewish roots or their earliest ancestors have names
associated with Ashkenazim. Four of
these have responded to private inquiries and all four who responded indicated
that their lines have traditions of being Cohanim. It is not yet clear whether there are any
other Cohanim within Haplogroup J2b.
However, if our suggestion of a relatively recent founding event for J2b1a-DYS455=8 is correct, there may be
other Cohanim lines found within the J2b1-M241 background in which the deletion
event in DYS455 occurred.
It is
also possible that non-Jewish members of the new subclade may eventually be
identified. Additionally, with a larger
sample size, we might locate Ashkenazim within the new subclade without a
tradition of being Cohanim. We cannot
rule out these possibilities because of the small number of members of the
subclade identified so far, and the possibility of selection bias. In future studies of Ashkenazi Jews, of
Cohanim, or of Haplogroup J2b, it would be very helpful if DYS455 were tested, at least in those
subjects who are members of Haplogroup J2, so that these questions could be
investigated with sufficient numbers to make firmer conclusions.
Y-STR Values of J2b-DYS455=8
Table
2 shows the Y-STR values for the 16 subjects. The first eight subjects are in J2b-M102 and
the last eight are also members of the new subclade of J2b-M241. Note that there is limited diversity among
the members of the new subclade, implying that it is fairly young.

None of
the Subjects 109-112 knows of a genealogical connection to any of the others,
and all four have different surnames.
Using Subject 112 as a point of comparison, the
time-to-the-most-recent-common-ancestor (TMRCA) calculator used by FTDNA,
called “FTDNATiP,” on the other two subjects who have 37 markers reported
(Subjects 109 and 110) results in a 50% probability of a TMRCA at 3 and 6
generations, respectively. Using only
the 25 markers in common to Subjects 112 and 111, the calculator provides a corresponding
50% TMRCA at 12 generations. Clearly,
this is an inadequate sample for precise determination of when the most recent
common ancestor lived, but it is also clear that it is very likely that this
ancestor lived in the last millennium.
Discussion
The
present study has identified a new subclade of Haplogroup J2b and the subclade
has been shown further to be a subgroup of Haplogroup J2b1a-M241. Precise placement of the new subclade with
respect to the previously known subgroups of J2b1a-M241 must await the
development of tests for their defining BPs and the identification of subjects
who are positive for each of them.
Only
about a dozen members of the new subclade have been identified so far, and only
about half of these have responded to inquiries, but all who responded have
indicated Ashkenazi and Cohanim backgrounds for their paternal lines. The geographic origin of the paternal lines,
where known, was most often indicated as Eastern Europe.
The addition of testing of DYS455 in future Jewish DNA studies would be useful, at least
for those subjects who are in Haplogroup J2 (i.e., positive for M172, which is
almost always tested).
As more
information is accumulated, the new subclade may be found to include non-Jewish
members. However, with the apparently
young age of the subclade—only several hundred years—it is also possible that
the deletion event in DYS455 occurred within an Ashkenazi community in Eastern Europe.
If this scenario is correct, then it implies that the deletion occurred
in an Ashkenazi Jew who was M241+ and whose father had a value of 11 on DYS455. There could be other Ashkenazi Jewish
lineages that have survived to the present from this ancestral M241+/455del-
background. All of those in the present
study who were M241+ and 455del-, reported that they were of northwest European
ancestry and had no Jewish roots, so it appears likely that M241+ became
established prior to the first M241+ persons becoming identified as Jewish.
Semino
(2004) found that 1.2% of his Ashkenazi subjects were M102+, which represents
about 5% of Ashkenazi J2’s. The new
subclade is too small to include all of these J2b’s. An interesting question for future studies
is, where are the other J2b Ashkenazim in the J2b phylogeny? Some are probably in J2b-M241*, but we do not
know at present if there are significant numbers in J2b-M205.
Web Resources
http://www.ysearch.org genetic
genealogy database
http://www.ybase.org genetic genealogy database
http://www.smgf.org genetic
genealogy database
http://home.comcast.net/~hapest5/index.html haplogroup predictor
References
Athey TW (2005) Haplogroup prediction using an
allele-frequency approach. J Genet Geneal, 1:1-7.
Athey TW (2006) Haplogroup prediction from Y-STR values using
a Bayesian-allele-frequency approach. J
Genet Geneal, 2:34-39.
Behar
DM, Thomas MG, Skorecki K, Hammer MF, Bulygina E, Rosengarten D, Jones
AL, Held K, Moses V, Goldstein D, Bradman N, Weale ME (2003) Multiple origins of Ashkenazi Levites: Y
chromosome evidence for both Near Eastern and European ancestries. Am J Hum
Genet 73:768–779.
Di
Giacomo F, Luca F, Popa LO,
Akar N, Anagnou N, Banyko J, Brdicka R, Barbujani G, Papola F, Ciavarella G, Cucci F, Di Stasi L, Gavrila
L, Kerimova MG, Kovatchev
D, Kozlov AI, Loutradis A, Mandarino V, C. Mammi C, Michalodimitrakis EN, Paoli G, Pappa
KI, Pedicini G, Terrenato
I, Tofanelli S, Malaspina
P, Novelletto A (2004) Y chromosomal haplogroup J as a signature of
the post-Neolithic colonization of Europe. Hum Genet 115:357-71.
ISOGG--International
Society of Genetic Genealogy (2007) Y Haplogroup Tree. Web Site URL:
http://www.isogg.org/tree/index.html.
King
RJ, Ozcan SS, Carter T, Kalfoğlu
E, Atasoy S, Triantaphyllidis
C, Kouvatsi A, Lin AA, Chow CE, Zhivotovsky LA, Michalodimitrakis M, Underhill PA (2008) Differential
Y-chromosome Anatolian influences on the Greek and Cretan Neolithic. Ann Hum
Genet. 72:205-214
.
Rootsi S, Magri C, Kivisild T, Benuzzi G, Help H, Bermisheva M, Kutuev I, Barac L, Pericic M, Balanovsky O, Pshenichnov A, Dion D, Grobei M, Zhivotovsky LA,
Battaglia V, Achilli
A, Al-Zahery
N, Parik J, King R, Cinnioglu
C, Khusnutdinova E, Rudan
P, Balanovska E, Scheffrahn
W, Simonescu M, Brehm A, Goncalves R, Rosa A, Moisan JP, Chaventre A, Ferak V, Furedi S, Oefner PJ, Shen P,
Beckman L, Mikerezi I, Terzic
R, Primorac D, Cambon-Thomsen
A, Krumina A, Torroni A,
Underhill PA, Santachiara-Benerecetti AS, Villems R, Semino O (2004) Phylogeography of
Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in
Europe. Am
J Hum Genet 75:128-137.
Semino O, Magri C, Benuzzi G, Lin A, Al-Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner
PJ, Zhivotovsky LA, King R, Torroni A, Cavalli-Sforza
LL, Underhill PA, A Santachiara-Benerecetti S
(2004) Origin, diffusion, and
differentiation of Y-Chromosome haplogroups E and J: Inferences on the neolithization of Europe and later migratory events in the
Mediterranean area. Am
J Hum Genet 74:1023-1034.
Sengupta
S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CT, Lin AA, Mitra M, Sil
SK, Ramesh A, Rani MVU, Thakur CM, Cavalli-Sforza LL, Majumder PP, Underhill PA
(2006) Polarity and temporality of high
resolution Y-chromosome distribution in India identify both indigenous and
exogenous expansions and reveal minor genetic influence of central Asian
pastoralists. Am
J Hum Genet , 78:202-221.




